Texas A&M AgriLife Research Study To Focus On Amino-Acid Radicals

Texas A&M AgriLife Research was recently awarded a grant by from the U.S. Department of Health and Human Services to study the function of amino-acid radicals, which are fundamental to both beneficial and harmful chemical reactions in living organisms.
A better understanding of these chemical reactions is likely to impact widespread disciplines from anticancer drug development to sustainable solar energy production to explaining how non-steroidal anti-inflammatory drugs, or NSAIDs, such as aspirin and ibuprofen function in the body. However, amino-acid radicals have been notoriously difficult to study.
“It is extremely challenging to experimentally resolve the thermodynamic and kinetic redox properties of a single amino-acid residue,” said Cecilia Tommos, professor in the Department of Biochemistry and Biophysics at Texas A&M. “Additionally, amino-acid radicals are typically highly oxidizing and reactive species, which increases the barrier for experiential characterization even further.”
To address these challenges, Tommos’ research group has developed a library of well-structured model proteins that will enable detailed studies of the fundamental chemical properties associated with redox-active amino acids.
Tommos is the primary investigator for the study, which will be conducted in collaboration with other research groups at Yale University, California Institute of Technology and Uppsala University in Sweden. Funding of $303,000 annually will be provided for the four-year grant, making the total in excess of $1.2 million.
Amino-Acid Radicals And The Enzymes That Use Them
Enzymes that utilize amino-acid radicals are called redox proteins. These enzymes are essential in the function of numerous metabolic pathways in plants, animals and various microorganisms, Tommos said. Chemically speaking, redox proteins and amino-acid radicals enable a wide range of biochemical processes including energy transformation, signal transduction and DNA replication and repair.
Redox proteins use metallocofactors, organic molecules and four types of amino acids — tyrosine, tryptophan, cysteine and glycine — to perform electron transfer, ET, and proton-coupled electron transfer, PCET, reactions.
“The four amino acids serve as essential one-electron, or radical, redox cofactors in biocatalytic and multistep ET/PCET processes — some of which are essential to life on earth,” Tommos explained.
However, she noted there is also a “sinister side” to these electron transfer reactions. When these reactions are induced by oxidative stress, they can cause significant cellular damage.
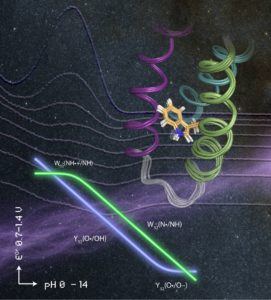
A Model System To Study Tyrosine And Tryptophan Radicals
Tommos said the family of well-structured model proteins her lab developed are specifically designed to study tyrosine and tryptophan radical formation, transfer and decay.
“This model system allows us to study amino-acid radical reactions under fully reversible conditions inside a structurally well-determined protein environment,” Tommos said.
She said the model proteins, named a3X proteins, were created by combining protein design with site-specific incorporation of unnatural or non-canonical amino acids and detailed structural studies.
“A major breakthrough with the a3X model system approach was made by the development of a very high potential protein film voltammetry method,” she said.
Protein film voltammetry is one of the techniques suitable for studying proteins that have electron transfer reactions, and is one method for studying the rates of enzyme-catalyzed chemical reactions.
Tommos said the unique properties of the a3X proteins, combined with protein film voltammetry, allowed her research group to measure reversible reduction potentials for redox-active tyrosine and tryptophan residues for the very first time.
“These results also provided us with an essential stepping-stone for what we wanted to accomplish with our next study,” she said.
Three Specific Aims
Tommos said the three specific aims for the current study will be to:
- Develop mechanistic and theoretical tools to study tyrosine/tryptophan-based multistep ET/PCET in model proteins and in natural enzymes.
- Expand the model protein system and measure thermodynamic and kinetic parameters associated with tyrosine/tryptophan-based multistep ET/PCET processes.
- Connect tyrosine/tryptophan oxidation and radical decay mechanisms to the dynamic properties of proteins.
“Each of these aims stand on their own and will provide critical first-of-their-kind information,” Tommos said. “If successfully conducted, these specific aims will result in a major advancement in our understanding of amino-acid radicals that, together with metals and organic redox cofactors, form the foundation for biological ET and PCET.”
This article by Paul Schattenberg originally appeared on AgriLife Today.